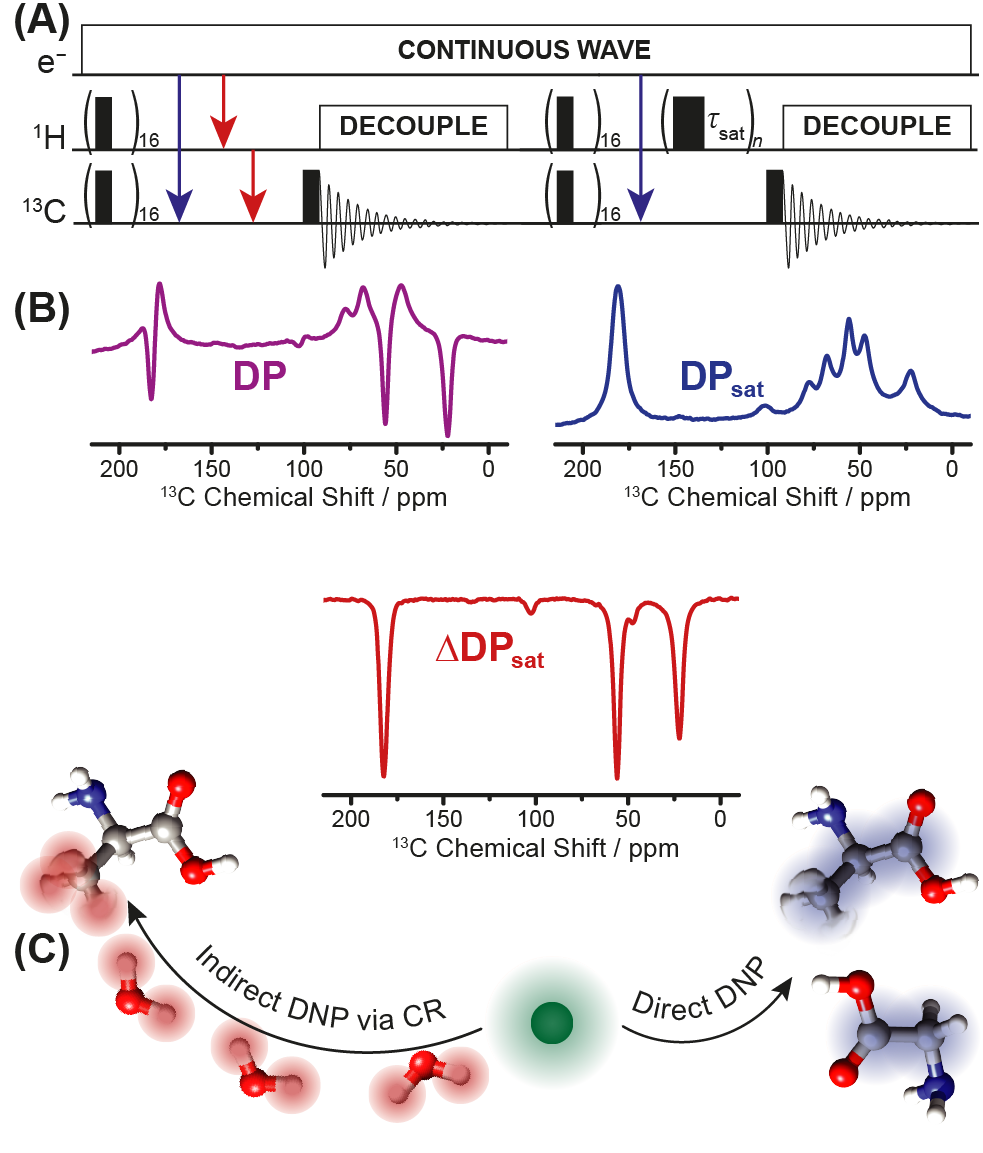
The concept of SCREAM-DNP (Specific Cross Relaxation Enhancement by Active Motions)
NMR is a powerful method for structure determination, unfortunately, it suffers from an intrinsic low sensitivity due to low polarization. This fact leads to the problem that the signal intensity often is not sufficient enough to resolve the investigated structure. The method of dynamic nuclear polarization (DNP) allows inscreasing the sensitivity within several orders of magnitude. But even with DNP the structural investigation of biomolecules can be very challenging, especially when it comes to structural determination in the natural environment, as it is necessary to be able to differentiate between the biomolecule of interest and other cellular components.
To solve this problem our group is using a new method the so-called SCREAM-DNP (Specific Cross Relaxation Enhancement by Active Motions). In this method, we use methyl-carrying “spy-molecules” that can specifically polarize biomolecules if those are in close proximity to the molecule interest. This can be achieved if the methyl-carrying molecule is specifically bound by the protein- or RNA-molecule of interest. We could show, that this method can be used for proof of binding between the “spy-molecule” and the biomolecule, moreover SCREAM-DNP can also serve as a spectral filter for components that do not interact with the spy-molecule.
Operating Principle
In general, the polarization transfer from the electron spin to the nuclear spin can occur either through direct or indirect polarization transfer. In the case of direct polarization (Figure (A) blue arrows) the polarization is directly transferred from the electron to the nuclei of interest (in this example to the carbon nuclei). In the case of indirect polarization, the transfer happens with an intermediate step, where the polarization is first transferred to protons and only afterward to the nuclei of interest (Figure (A) red arrows). If the sample contains mobile methyl-groups, both polarization pathways can occur simultaneously. This effect is caused by the very fast reorientation dynamics of methyl-groups (Nuclear Overhauser Effect). In such a case, the direct pathway results in positive and indirect transfer in negative signals. This means that the resulting spectra contain negative as well as positive spectral intensities (Figure (B),DP, lilac). To separate the negative and positive compound of the spectra and isolate just the information caused by the methyl-groups, which would allow the investigation of the near surrounding of the methyl-group, another experiment has to be performed where the use of saturation pulses (Figure (A) τsat ) allows to suppressing the indirect pathway through protons. The resulting spectra contain only positive signals caused by direct polarization (Figure (B), DPsat, blue). Mathematical subtraction of DP and DPsat leads the spectral contribution from the indirect pathway caused due to the fast reorientation dynamics of methyl groups (Figure (B), ΔDPsat ). This spectrum than contains information about the surrounding of the methyl-groups. Figure (C) shows schematic representation of the direct and indirect pathway on the example of glycine, which does not have any methyl-groups and alanine, which has exactly one methyl group and that shows significant SCREAM-activity.