DNP in biomolecular complexes
Dynamic nuclear polarization (DNP) in biomolecular complexes can be very difficult. As DNP is typically performed at cryogenic temperatures, typically around 100 K, the investigated biomolecules have to be cryoprotected from degradation. This is mostly performed by adding glycerol to the sample, whereas it must be ensured that glycerol does not affect the correct folding or the function of the molecule. Moreover, the complexity of biomolecular NMR lays in the spectral signal overlapping as those contain similar structural building blocks such as amino acids in the case of proteins or nucleotides in the case of nucleic acids. Therefore our group focuses on DNP-methods that allow systematic investigation of specific sides in the biomolecule.
Ribonucleic acids (RNA)
Ribonucleic acids (RNA) are not only essential for the translation of the genetic code into proteins, but they are more fulfill a multitude of tasks and functions in our cells. Despite the fact, that RNA constructed just by four building blocks, the nucleotides adenosine, guanosine, cytosine and uridine, the variety of RNA is enormous. In general, RNA can be distinguished in coding and non-coding RNA (ncRNA). While the coding messenger RNA (mRNA) is translated into the protein the ncRNAs perform different regulatory tasks in the cell. Some prominent examples for ncRNA are the ribosomal RNA (rRNA) and the transfer RNA (tRNA), which are needed for gene expression, the micro RNA (miRNA) that plays an important role in the chromatin folding, the small interfering RNA (si RNA) which is necessary for the histone modification, the small nuclear (snRNA) that is involved in the process of splicing in eukaryotic organisms or the so-called small nucleolar RNA (snoRNA), that is crucial for the processing and modification of other ribonucleic acids. The ncRNAs are often found in a complex with proteins, the so called ribonucleoproteins (RNPs). Another type of RNAs are the RNA-aptamers, which are mostly selected by the ability to bind a very specific ligand with high affinity. Those are interesting for applications in the analytical sciences or cancer research.
The tetracycline binding aptamer (TC-aptamer)
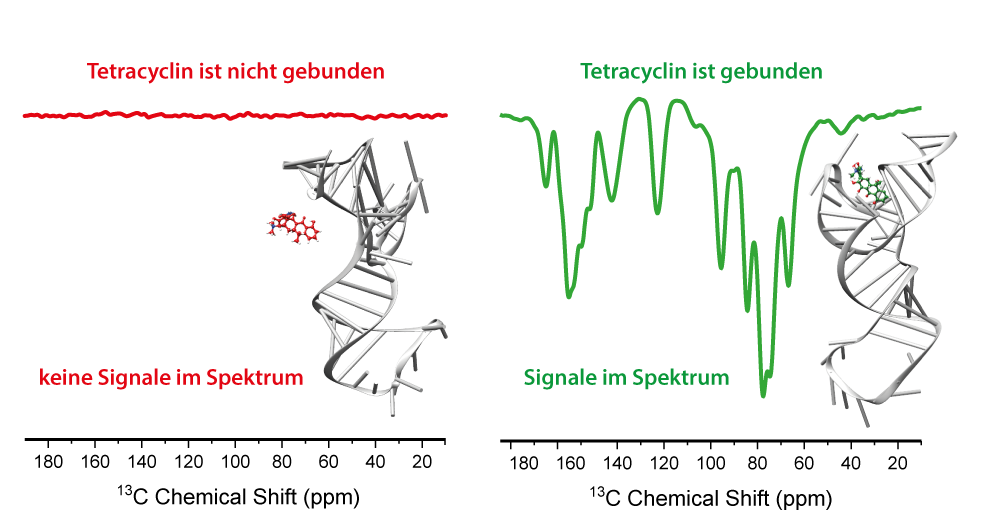
The TC-aptamer was originally found to investigate the interaction between RNA and the antibiotic tetracycline. But after several modifications, it could be used for the regulation of gene expression or splicing (Suess et al., Nucleic Acids Res. 2003, 31,1853). The antibiotic tetracycline contains three methyl-groups and is bound with a very high affinity by the RNA. As the RNA in its natural form does not contain any methyl groups the TC-aptamer was a perfect system to investigate SCREAM-DNP (see chapter SCREAM-DNP). To do so tetracycline was labeled with NRM active carbon-nuclei (13C) and was used as a methyl-carrying spy molecule. If tetracycline was bound to the RNA characteristic spectral fingerprint of the RNA-aptamer occurred, which could be isolated from the unwanted background signal. By changing the experimental conditions and preventing binding between the RNA and the tetracycline, the specific signals do not occur.
This study was published in 2019 in the Journal Angewandten Chemie (Aladin et al., Angew. Chem. Int. Ed. 2019, 58, 4863), for which Victoria Aladin got the GDCh-Ernst-Award. This study demonstrates the first application of SCREAM-DNP for selective investigation in biomolecular complexes. Because of the very high specificity of this method the complexes could also be investigated beside other components in the sample, as the spectral components can be spectrally isolated from the unwanted background.
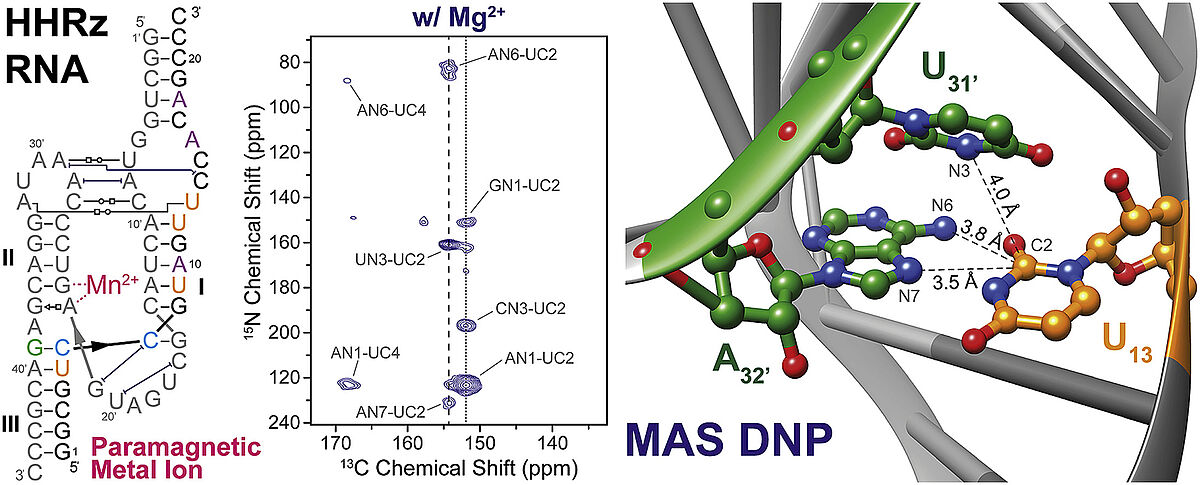
The hammerhead-ribozyme (HHRz)
The hammerhead-ribozyme (HHRz) is a structural component of the viral genome in plants. After the successful infection of the host cell, the HHRz catalyzes an enzymatic cleavage reaction, which allows the replication of viral RNA. For this cleavage a specific binding site has to be occupied with divalent ions such as Mg2+ or Mn2+. For a long time, it was unknown if the coordination of the divalent ions entails significant structural change until Martick and Scott showed that the coordination of the divalent ion does not affect the crystal structure of the HHRz (Scott et al., Prog. Mol. Biol. Transl. Sci., 2013, 120, 1). Our group could confirm this hypothesis by using DNP enhanced MAS-NMR. As well in the presence as in the absence of Mg2+ the 2D-correlation spectra (15N-13C-TEDOR) show the same tertial contacts between two RNA stem-loops by non-canonical reverse Hoogsteen-basepair. This strongly suggests, that also in solution the specific coordination of Mg2+ does not significantly affect the structural population. (Daube et al., Solid State Nucl. Magn. Reson. 2019, 101, 21).
Moreover, we could show that Mn2+ could be utilized as polarizing agent for performing site specific DNP. This is highly interesting as it shows that an exogenous polarizing agent, such as nitroxide radical, does not have to be added to the sample solution, as DNP is performed by using the bound metal ion inside the biomolecule as polarizing agent (Wenk et al., J. Biomol. NMR 2015, 63, 97).